Note
Click here to download the full example code
Learning an SPN for Classification
We can perform classification by learning an SPN from data and then comparing the probabilities for the given classes.
import numpy as np
np.random.seed(123)
from spn.algorithms.LearningWrappers import learn_parametric, learn_classifier
from spn.structure.leaves.parametric.Parametric import Categorical, Gaussian
from spn.structure.Base import Context
import matplotlib.pyplot as plt
import seaborn as sns
Out:
/opt/hostedtoolcache/Python/3.6.15/x64/lib/python3.6/importlib/_bootstrap.py:219: RuntimeWarning: numpy.ufunc size changed, may indicate binary incompatibility. Expected 192 from C header, got 216 from PyObject
return f(*args, **kwds)
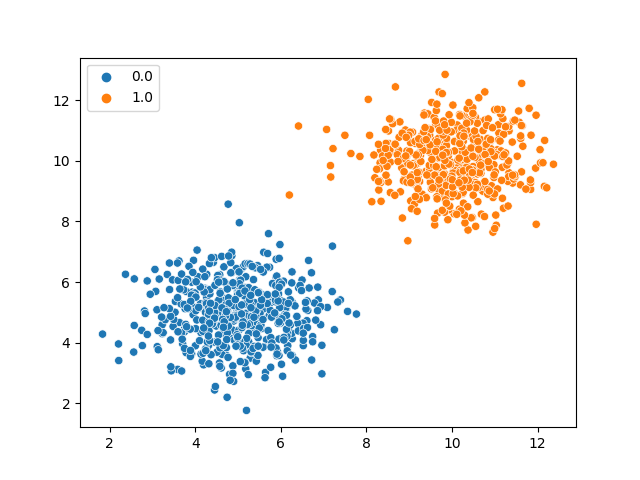
Imagine we have the following dataset generated by two gaussians with means \((5,5)\) and \((10,10)\), and we label the cluster at \((5,5)\) to be class 0 and the cluster at \((10,10)\) to be class 1.
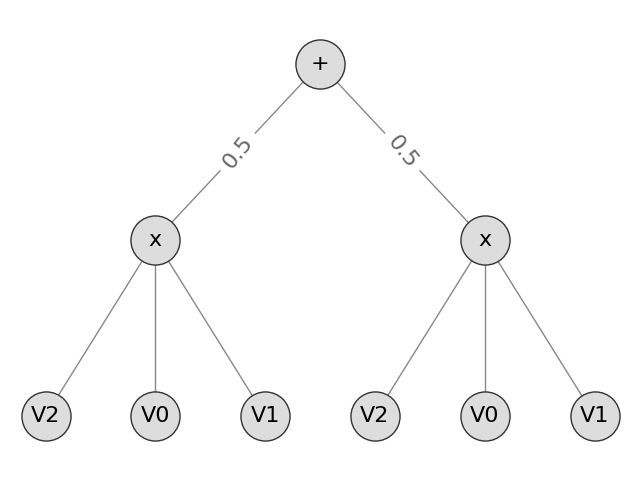
Here, we model our problem as containing 3 features: two Gaussians for the coordinates and one Categorical for the label. We specify that the label is in column 2, and create the corresponding SPN.
train_data = np.c_[
np.r_[np.random.normal(5, 1, (500, 2)), np.random.normal(10, 1, (500, 2))],
np.r_[np.zeros((500, 1)), np.ones((500, 1))],
]
sns.scatterplot(train_data[:, 0], train_data[:, 1], hue=train_data[:, 2])
Out:
/opt/hostedtoolcache/Python/3.6.15/x64/lib/python3.6/site-packages/seaborn/_decorators.py:43: FutureWarning: Pass the following variables as keyword args: x, y. From version 0.12, the only valid positional argument will be `data`, and passing other arguments without an explicit keyword will result in an error or misinterpretation.
FutureWarning
<AxesSubplot:>
We can learn an SPN from the training data:
spn_classification = learn_classifier(
train_data, Context(parametric_types=[Gaussian, Gaussian, Categorical]).add_domains(train_data), learn_parametric, 2
)
from spn.io.Graphics import draw_spn
draw_spn(spn_classification)
Out:
/opt/hostedtoolcache/Python/3.6.15/x64/lib/python3.6/importlib/_bootstrap.py:219: RuntimeWarning: numpy.ufunc size changed, may indicate binary incompatibility. Expected 192 from C header, got 216 from PyObject
return f(*args, **kwds)
<module 'matplotlib.pyplot' from '/opt/hostedtoolcache/Python/3.6.15/x64/lib/python3.6/site-packages/matplotlib/pyplot.py'>
Now, imagine we want to classify two instances, one located at \((3,4)\)
and another one at \((12,8)\). To do that, we first create an array with
two rows and 3 columns. We set the last column to np.nan to indicate
that we don’t know the labels. And we set the rest of the values in the 2D
array accordingly.
We can do classification via approximate most probable explanation (MPE). Here, we expect the first instance to be labeled as 0 and the second one as 1.
from spn.algorithms.MPE import mpe
print(mpe(spn_classification, test_data))
Out:
/home/runner/work/SPFlow/SPFlow/src/spn/structure/leaves/parametric/Inference.py:88: RuntimeWarning: divide by zero encountered in log
probs[idx_in] = np.array(np.log(node.p))[cat_data[~out_domain_ids]]
[[ 3. 4. 0.]
[12. 18. 1.]]
Total running time of the script: ( 0 minutes 0.883 seconds)